|
|
|
|
| Resistance |
|
| Resistance: Introduction |
| HIV resistance � the basics |
| Clinical implications of drug resistance |
| How can HIV resistance be avoided? |
| HIV resistance tests |
| What strategies are being investigated to help overcome drug resistance? |
| References |
|
|
Resistance: Introduction
Current treatment regimens for HIV infection generally employ combinations of three or more potent antiretroviral drugs, which in many cases can drive down the level of virus in the body to undetectable levels. However, this approach is not always successful. A significant number of individuals have persistently high levels of HIV despite potent medication, while others experience only transient virological suppression before their viral load rapidly rebounds to near pre-treatment levels.
One of the major causes of treatment failure is HIV drug resistance � the "Achilles� heel" of antiretroviral therapy. This section explains what causes HIV resistance, how it can be monitored, and what steps can be taken to minimise its potential impact on successful treatment.
|
|
|
HIV resistance � The basics
What is resistance?
- Antiretroviral drugs were first introduced in the late 1980s. They were developed to target �wild-type� HIV � the most common form or �strain� of the virus � and work by blocking critical viral enzymes required for the replication of functional virus.
- Soon after these drugs were introduced, it became apparent that virus was somehow able to escape the action of antiretrovirals, leading to a loss of response to medication. This evasion of the inhibitory action of antiretrovirals is called HIV drug resistance.
What causes drug resistance to develop?
- HIV becomes resistant to the action of a drug as a result of one or more mutations in the viral genome � aberrations in the genetic code of the virus. These aberrations result in the production of viral proteins that are subtly different to their wild-type counterparts in structure or functioning, so that although they are still able to play their role in HIV replication, they are not targeted as effectively as wild-type by antiretroviral agents.
- Mutations occur spontaneously in all living organisms. The frequency of mutation depends partly on how often the organism reproduces or its cells divide. Fast-growing viruses like HIV replicate millions of times a day, so mutations are constantly arising.
- Drug therapy does not cause the virus to develop drug-resistant mutations
� they occur naturally and spontaneously in the absence of treatment,1�3 as HIV has a high error rate when replicating its genetic code (RNA).4 The combination of a high error rate and rapid pace of HIV replication5,6 ensures that these drug-resistant mutants are consistently present within the genetically diverse HIV population, termed a quasispecies.
- It has been estimated that in uncontrolled HIV infection, every possible single mutation in the HIV genome could arise many times in a patient�s viral population each day (Coffin 1995).
- However, in the absence of drug therapy these naturally occurring mutants are inferior in their ability to replicate and compete compared with the dominant wild-type viruses (i.e. they are less �fit�), and therefore exist at only low levels.7,8
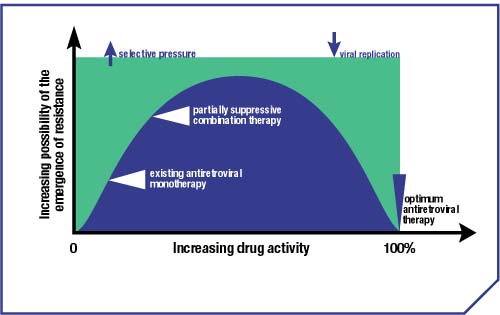
Figure 1. Relationship between drug activity and the emergence of drug-resistant mutants
- The initiation of drug therapy exerts a strong, selective pressure on the HIV population. Wild-type virus is targeted and prevented from replicating effectively, whereas a mutant virus strain that is able replicate in the presence of drug will thrive, and come to dominate the viral population (following the principles of natural selection and �survival of the fittest�) (Figures 1 & 2).9,10
- It is important to remember that the emergence of drug-resistant HIV is a dynamic process. Distinct viral strains, with differing sensitivities to antiretroviral drugs, can co-exist in an infected person, albeit at different levels. Even if drug therapy is stopped, drug-resistant virus may continue to persist, although over time wild-type virus is likely to predominate in the viral population.11�13
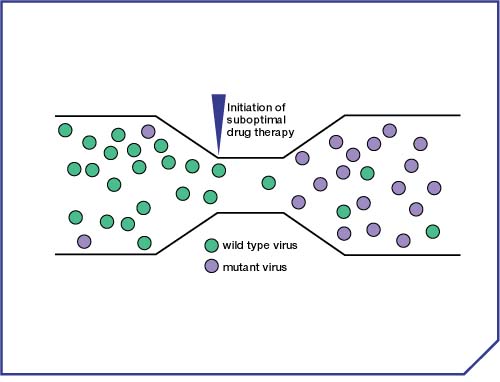
Figure 2. Selection of resistant virus under drug pressure
Breaking the mutations code
- The wild-type HIV genome, which carries the genetic code of the virus, is made up of a specific sequence of nucleotide bases, which codes for the various viral proteins. Genetic mutations change the amino acid sequences of the proteins they code for, for example by substituting one type of amino acid for another, so they are named according to their position in the protein and the amino acid alteration they encode.
- For example:
- the M184V mutation associated with lamivudine resistance refers to a substitution at position 184 (�codon� 184) of the HIV reverse transcriptase (RT) enzyme : the amino acid methionine (M), which normally occurs in wild-type virus, has been substituted by valine (V)14
- the T69S-S-S insertion mutation, associated with resistance to multiple nucleoside analogue RT inhibitors (NRTIs), indicates codon 69 has changed from a threonine (T) to a serine (S) and that two additional serine (S) amino acids have been inserted between positions 69 and 70 of RT.15�18
Primary mutations are those mutations that interfere with the inhibitory action of the drug and result in an increase in the amount of drug necessary to inhibit viral replication.
Secondary mutations increase the level of resistance by improving the fitness of viruses carrying primary mutations. They usually do not render the virus resistant to drug in the absence of primary mutations.
How is drug resistance measured?
- The level of drug resistance is assessed in the lab (�in vitro�), by comparing the susceptibility of patient virus samples to the inhibitory effects of a specified drug with the susceptibility of wild-type virus to the same drug. This is done by measuring the concentration of drug required to reduce replication of both the patient sample and the wild-type control by a fixed amount, typically by 50% (IC50) or 90% (IC90) (Figure 3).
- The IC50 or IC90 for the patient isolate is divided by that of the wild-type control and the difference expressed as an �x-fold reduction in susceptibility� or �x-fold resistance�. For example, a viral isolate from an infected person with an IC50 twice as high as the wild-type is said to show 2-fold resistance/reduced susceptibility to that drug.
- Drug inhibitory concentrations can also be measured in HIV-infected people (ie. �in vivo�) by determining the level of a drug in blood plasma that reduces the plasma viral load by a fixed amount. These in vivo inhibitory concentrations are termed �effective concentrations� (EC) and, for example, an EC50 is the plasma concentration of a drug that halves the viral load, while the EC90 is the concentration that reduces it to 10% of its original level. Effective concentrations are not used to determine an isolate�s level of resistance, but are valuable for determining what level of drug in the body will suppress a resistant viral strain by a given amount.
- This provides a clue as to the importance of maintaining high drug levels in the body: if drug levels are reduced, replication of both wild-type and mutant viruses is less likely to be inhibited, allowing the emergence of further mutations that increase the level of resistance.
Figure 3.
In vitro antiviral potency � effect of resistance
Top
|
|
|
Clinical implications of drug resistance
Why combination therapy?
- The number of mutations required for resistance to a drug is clearly an important factor in determining the rate at which drug resistance will emerge. The chances of a single mutation occurring randomly in the population are relatively high, whereas the likelihood of several specific mutations occurring at any one time in the same genome is significantly less.
- Thus, resistance to drugs such as lamivudine and nevirapine, which can be rendered ineffective by a single mutation in the viral genome, emerges within weeks of initiating monotherapy.19,20 By contrast, resistance to zidovudine is typically a consequence of the stepwise accumulation of multiple mutations, and is, therefore, slower to develop under the same circumstances.21
- This underpins the rationale behind combination antiretroviral therapy. If HIV is simultaneously targeted in several different ways, the probability of the emergence of a fully functional mutant strain that is capable of rendering the combined therapy ineffective is much lower (Figure 4). And once the level of virus has been driven down by potent therapy, the opportunity for such a mutant to emerge will be dramatically reduced, in line with the low number of replicating viruses in the body.
Figure 4. Effective HAART reduces the probability of functional, fully resistant virus emerging
What is cross-resistance?
- Because drugs of the same type or �class� (e.g. protease inhibitors) have a similar mechanism of effect, it is inevitable that in some instances a mutation or pattern of mutations (genotype) that confers resistance to one drug will also affect the susceptibility of the virus to another in its class. This is termed cross-resistance.
- For example, the protease inhibitors ritonavir and indinavir share a similar resistance profile (Figure 5). Mutations at codon 82 of the protease gene (V82A, V82T or V82F) seem to play a central role in the development of resistance to both drugs, and many of the same mutations are apparent in ritonavir-resistant virus and indinavir-resistant virus.
- For protease inhibitors, the degree of cross-resistance appears to depend not only on the type of mutation, but also on the number of mutations accumulated � which in turn is associated with the length of time that mutant virus is exposed to the same drug regimen. Continued viral replication on a failing regimen allows the emergence of more complex patterns of mutations and cross-resistance under multidrug selection pressure, as well as the evolution of resistance to all the drugs in the regimen. Early failure on any anti-HIV regimen (with or without protease inhibitors) is generally characterised by simple patterns of resistance to only one or two drugs. In some cases, where low drug levels have allowed the viral load to increase rather than the presence of drug-resistant variants, early failure can be characterised by a wild-type viral genome. However, continued replication while drugs are still being taken will allow resistance to develop.
Figure 5. Protease gene mutations selected by PIs
Multidrug resistance
- Some genotypes have been identified that result in resistance across an entire class of drugs. For example, the Q151M mutation in reverse transcriptase confers partial resistance to all currently approved NRTIs � termed multinucleoside resistance. Accumulation of secondary mutations at four additional sites serves to increase the level of resistance to this class of drugs as well as improve the level of fitness of the Q151M mutant strain.22
- Another multidrug resistance genotype is the K103N NNRTI-associated mutation, which confers resistance to all of the currrently available non-nucleoside RT inhibitors.23 This mutation is the most common to emerge on failure of efavirenz,24 and is also commonly associated with delavirdine25 and nevirapine19. However, although K103N is the most well-recognised NNRTI cross-resistance mutation, the NNRTIs are a highly cross-resistant class, and failure of any one with the emergence of NNRTI-associated mutations generally rules out the use of any other.
- As they have different modes of action, there is no cross-resistance between different classes of antiretroviral drug e.g. between NRTIs and NNRTIs. However, it is entirely feasible for a virus to carry mutations that result in resistance to more than one class of drug. Indeed, some virus strains have been identified that have reduced susceptibility to all three currently approved classes of drug.26
Top
|
|
|
How can HIV resistance be avoided?
A primary goal of antiretroviral therapy is maximal and durable suppression of viral load � to keep the level of virus as low as possible for as long as possible.27 By minimising the reservoir of replicating virus in the body, the chances of mutant virus being generated are severely reduced. Viral load levels between 50 and 500 copies/ml are associated with a higher risk of resistance than levels below 50 copies/ml.28
This is best achieved with a potent combination of three or more antiretroviral drugs in a therapeutic strategy known as �highly active antiretroviral therapy� (HAART), which together impose a high �genetic barrier� to resistance. An accumulation of multiple mutations in the viral genome is then needed if the virus is to evade the effects of all drugs in the regimen. However, the sensitivity of modern viral load assays can often detect the increase in viral replication associated with the emergence of resistance to even one drug in the combination. As a result, therapeutic failure can be defined before all the drugs in the regimen have been rendered ineffective, significantly improving the prospects for success on a new regimen.
This approach has the greatest chance of succeeding if inhibitory drug levels are consistently maintained, which requires good adherence to the treatment regimen. If drug doses are missed, or are not taken in accordance with dosing instructions (e.g. with/without food, etc.), this will adversely affect the level of drug in the body and provide a better opportunity for drug-resistant virus to emerge.
Regular viral load monitoring is recommended (every 2�4 months),27,29 as an increase in viral levels may indicate the emergence of resistant virus. If this is the case, switching to a new antiretroviral regimen at an early stage may limit the development of additional secondary mutations that may adversely affect the response to subsequent therapies.
However, it is important to realise that there are several other potential reasons for treatment failure, including concomitant illness affecting drug availability in the body, drug interactions reducing blood levels of the antiretroviral and gastrointestinal intolerance, also leading to low drug levels in the blood.
Rational Treatment Sequencing
- Even in individuals who at the outset respond well to drug treatment, viral load may eventually rebound and a new treatment regimen may need to be considered (Figure 6). A long-term treatment plan is, therefore, likely to involve a sequence of different antiretroviral drug combinations.
Figure 6. Durability of response to triple therapy : 106 optimal responders to indinavir therapy (viral load < 500 copies/ml before week 24 and maintained for > 12 weeks).
Source: Holder DJ, Shivaprakash M, Danovich RM, et al. Duration of HIV-1 load suppression in patients treated with indinavir who experience virus load declines to <500 vRNA copies/ml. Antiv Ther 1997; 7:118.
- If therapeutic failure occurs as a result of drug resistance, it seems logical to switch to other drugs that the individual has not previously tried and to which HIV is unlikely to be resistant. Clinicians, therefore, need to think strategically in terms of drug sequencing, and, given the problem of cross-resistance, aim to preserve future treatment options, where possible.
- The ability to replace one failing drug with another from the same class (known as drug sequencing) is important for preserving the length of effective therapy. Drug sequencing has been attempted for the NNRTIs, but the results have generally been disappointing due to the cross-resistance problems mentioned above.30�32 Greater success with drug sequencing has been achieved with protease inhibitors.
- For example, resistance emerging to both nelfinavir and amprenavir shows low cross-resistance with other PIs,33,34 although amprenavir�s I50V primary mutation is associated with cross-resistance to ritonavir35 and appears to confer cross-resistance to lopinavir36,37 - the active component of Kaletra, which selects the I50V mutation in the laboratory.38
- However, the primary D30N mutation selected by nelfinavir is unique amongst the PIs, and the activity of other PIs against nelfinavir-resistant HIV has been shown to be preserved39 provided that nelfinavir is replaced before accumulating secondary mutations lead to cross-resistance developing.40
- Replacing a failing nelfinavir-containing drug regimen with a second regimen based on saquinavir and ritonavir has been found to give sustained suppression of HIV in clinical trials.41,42
- The results demonstrate that the principle of rational drug selection and sequencing can be applied to extend the length of effective therapy across treatment lines. Further research, and the introduction of new drugs and drug classes with patterns of resistance different to the current agents, can be expected to further improve the situation.
Transmission of drug-resistant virus
- With the increasing prevalence of drug-resistant virus, primary HIV resistance (where an individual is initially infected with HIV that is drug resistant) is increasingly common, and has been shown to be on the increase in both Europe and the USA.43�46
- Primary HIV resistance may increase the likelihood that an initial antiretroviral treatment combination will not be effective, as virus may already be resistant to one or more components of the regimen.
Top
|
|
|
HIV resistance tests
Two types of test for establishing the presence of drug-resistant HIV in an individual are available:
identify individual mutations in the HIV genome of patient-derived virus samples. These are compared with known resistance mutations for currently available antiretroviral drugs, and a conclusion is drawn as to the likelihood of virus having reduced susceptibility to those drugs.
Phenotype assays directly measure the ability of antiretroviral drugs to inhibit replication of patient-derived virus samples, by culturing virus in the laboratory in the presence of various concentrations of one or more drugs. The susceptibility of the patient�s virus is compared with that of a drug-susceptible control strain.
A number of trials and treatment guidelines indicate that HIV resistance tests of both types can aid in patient management by improving at least short-term virologic outcome when the results are used to help determine which drugs to use in a therapeutic combination after failure of a previous regimen.29,27,47�52 They can help to determine if drug resistance is the cause of treatment failure in an individual, and can give an indication of what treatment options may be open.
- Generally, studies have shown that the presence of drug susceptibility is predictive of treatment response and adds to knowledge obtained from either drug history or viral load measurements alone.42,53,54 However, a good response is not uniformly seen when virus is sensitive at baseline. This may reflect the presence of minority drug-resistant viral subpopulations or factors such as suboptimal drug levels and poor adherence.55
- Drug resistance tests are not the sole criterion for deciding when to initiate or change therapy. Other important factors, including patient's drug history, viral load, tolerance and adherence should be considered prior to switching treatment regimens.
How are resistance tests currently used?
- The following is a summary of current US (IAS�USA Panel) and Pan-European (EuroGuidelines Group) recommendations for the use of antiretroviral resistance testing.27,55,56
|
|
IAS�USA panel |
EGG |
|
Treatment-naive |
Primary infection
Consider testing
Established infection
Consider testing |
Primary recent infection
Recommend testing;
But consider:
Time since infection
Possibility of transmission
from a treated individual
Chronic infection
Consider testing
Post-exposure prophylaxis
Test index case if
available but do not wait
to initiate |
|
Treatment-experienced |
First regimen failure
Recommended testing
Multiple regimen failures
Recommended testing
Pregnancy
Recommended testing |
All treated patients
Recommended testing
Pregnancy
Recommended testing |
Important but as yet unresolved issues in drug resistance testing include:
- the optimum method(s) of interpreting test results to provide maximum benefit on a subsequent treatment regimen
- the long-term benefit of resistance testing in terms of virological outcome
- identification of situations in which one type of resistance testing is superior to the other
- whether resistance testing will be valuable for all drugs and drug classes
- development of more sensitive assays that can reliably test samples from individuals with a low viral load (< 500 copies/ml)
- the impact of mutational interactions, concomitant drug administration and the potential for cross-resistance on resistance levels for component drugs in the various multidrug combination regimens that are available
- the cost-effectiveness of resistance testing as a routine part of patient monitoring.
Top
|
|
|
|
What strategies are being investigated to help overcome drug resistance?
Clinicians and research scientists recognise the importance of drug resistance and its consequences, and this field is now at the forefront of HIV research. We are continually learning more about resistance and how it can be controlled to best effect. A number of potential strategies to help improve our ability to delay or elmininate resistance are currently under investigation:
- The development of new antiretrovirals with activity against HIV strains that are resistant to currently approved drugs:
- Drugs currently in development include new NRTIs and second generation PIs and NNRTIs, with novel resistance profiles, as well as agents offering a new line of attack on the virus, such as integrase inhibitors and entry inhibitors (e.g. fusion inhibitors and chemokine coreceptor inhibitors).
Protease inhibitor boosting (increased PI exposure by co-administration of a second drug e.g. ritonavir)
- When exposure to an effective antiviral drug is high, virus is most likely to be inhibited and thus antiviral resistance is least likely to occur.
- Higher plasma levels of protease inhibitor may increase the potential to suppress some resistant viral strains.57
- PI boosting helps to simplify treatment regimens, and thus is likely to have a positive impact on adherence rates.
Therapeutic drug monitoring
- The level of active drug that is achieved in the blood after standard dosing varies from one individual to another, and in some patients inadequate drug levels (due to inadequate absorption, rapid metabolism or drug interactions) may contribute to treatment failure and encourage the emergence of drug-resistant virus.58,59 There is growing interest in the possibility of using drug level monitoring to optimise antiretroviral therapy by increasing drug doses to address suboptimal drug levels in the blood.
Top
|
|
|
References
Mohri H, Singh MK, Ching WTW, et al. Quantitation of zidovudine-resistant human immunodeficiency virus type 1 in the blood of treated and untreated patients. Proc Natl Acad Sci USA 1993; 90:25�29.
N�jera I, Richman DD, Olivares I, et al. Natural occurrence of drug resistance mutations in the reverse transcriptase of human immunodeficiency virus type 1 isolates. AIDS Res Hum Retroviruses 1994; 10:1479�1488.
N�jera I, Holgu�n A, Qui�ones-Mateu E, et al. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol 1995; 69:23�31.
Mansky LM and Temin HM. Lower in vivo mutation rate of human immunodeficiency virus -type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 1995; 69:5087�5094.
Wei X, Ghosh SK, Taylor ME, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 1995; 373:117�22.
Ho DD, Neumann AU, Perelson AS, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 1995; 373:123�126.
Chow YK, Hirsch MS, Merrill DP, et al. Use of evolutionary limitations of HIV-1 multidrug resistance to optimize therapy. Nature 1993; 361:650�654.
Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis and therapy. Science 1995; 267:483�489.
Frost SDW, McLean AR. Quasispecies dynamics and the emergence of drug resistance during zidovudine therapy of HIV infection. AIDS 1994; 8:323�332.
Kellam P, Boucher CAB, Tijnagal JMGH, et al. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol 1994; 75:341�351.
Albert J, Wahlberg J, Lundeberg J, et al. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in post-treatment sera. J Virol 1992; 66:5627�5630.
Condra JH, Schleif WA, Blahy OM, et al. Evidence for the existence of long-lived genetic reservoirs of HIV-1 in infected patients. 4th International HIV drug-resistance Workshop, Sardinia, 1995; Abstract 82.
Devereux HL, Youle M, Johnson MA, et al. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 1999; 13:123�127.
Boucher CA, Cammack N, Schipper P, et al. High-level resistance to (-) enantiomeric 2'-deoxy-3'-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 1993; 37:2231�2234.
Bloor S, Hertogs K, Desmet RL, et al. Virological basis for HIV-1 resistance to stavudine investigated by analysis of clinical samples. Antivir Ther 1998; 3 (Suppl 1):13.
de Jong JJ, Goudsmit J, Lukashov VV, et al. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 1999; 13:75�80.
Whitcomb JM, Limoli K, Wrin T, et al. Phenotypic and genotypic analysis of stavudine-resistant isolates of HIV-1 [Abstract 17]. Antivir Ther 1998; 3 (Suppl 1):14.
Winters MA, Cooley KL, Girard YA, et al. Phenotypic and molecular analysis of HIV-1 isolates prossessing 6 bp inserts in the reverse transcriptase gene that confer resistance to nucleoside analogues [Abstract 16]. Antivir Ther 1998; 3 (Suppl 1):14.
Richman DD, Havlir D, Corbeil J, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol 1994; 68:1660�1666.
Schuurman R, Nijhuis M, van Leeuwen R, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J Infect Dis 1995; 171:1411�1419.
Boucher CAB, O'Sullivan E, Mulder JW, et al. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis 1992; 165:105�110.
Iversen AKN, Shafer RW, Wehrly K, et al. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol 1996; 70:1086�1090.
Bacheler L, Jeffrey S, Hanna G, et al. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol 2001; 75:4999�5008.
Bacheler LT, Anton ED, Kudish P, et al. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy Antimicrob Agents Chemother 2000; 44:2475�2484.
Demeter LM, Shafer RW, Meehan PM et al. Delavirdine susceptibilities and associated reverse transcriptase mutations in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdine monotherapy Antimicrob Agents Chemother 2000; 44:794�797.
Shafer RW, Winters MA, Palmer S, et al. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann Intern Med 1998; 128:906�911.
Guidelines for the Use of Antiretroviral Agents in HIV-Infected Adults and Adolescents, DHHS, March, 2004.
Raboud JM, Montaner JS, Conway B, et al. Suppression of plasma viral load below 20 copies/mL is required to achieve a long-term response to therapy. AIDS 1998; 12:1619�1624.
Carpenter CJ, Cooper DA, Fischl MA, et al. Antiretroviral Therapy in Adults: Updated Recommendations of the International AIDS Society�USA Panel. JAMA 2000; 283:381�390.
MacArthur RD, Kosmyna JM, Crane LR, et al. Sequencing of non-nucleoside reverse transcriptase inhibitors based on specific mutational patterns fails to lower plasma HIV-RNA levels in persons extensively pre-treated with antiretrovirals who are failing virologically on nevirapine-containing antiretroviral regimens. Seventh European Conference on Clinical Aspects and Treatment of HIV-Infection. Lisbon, Portugal, 23�27 October 1999. Abstract 208.
Shulman N, Zolopa A, Murlidharan U, et al. Efavirenz (EFV) and adefovir (ADV)-based salvage in antiretroviral experienced HIV+ patients. 39th Interscience Conference on Antimicrobial Agents and Chemotherapy. San Francisco, California, USA, 26 � 29 September 1999. Abstract 2201.
Zaccarelli M, Cingolani A, Forbici F, et al. Cross-resistance among NNRTIs: evaluation of the option of recycling efavirenz after nevirapine failure. Antivir Ther 2001; 6:S26.
Tisdale M, Myers RE, Maschera B, et al. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother 1995; 39:1704�1710.
Kemper CA, Witt MD, Keiser PH, et al. Sequencing of protease inhibitor therapy: insights from an analysis of HIV phenotypic resistance in patients failing protease inhibitors. AIDS 2001; 15:609- 615.
Snowden W, Shortino D, Klein A, et al. Development of amprenavir resistance in NRTI-experienced patients:alternative mechanisms and correlation with baseline resistance to concomitant NRTIs. Antivir Ther 2000; 5 (Suppl 3):84.
Parkin NT, Chappey C, Maranta M, et al. Genotypic and phenotypic analysis of a large database of patient samples reveals distinct patterns of protease inhibitor cross-resistance. Antivir Ther 2001; 6 (Suppl 1):49�50.
Prado JG, Wrin,T, Beauchaine J, et al. Lopinavir resistance of amprenavir-selected, replication-impaired mutants of HIV-1. Antivir Ther 2001; 6 (Suppl 1):51.
Mo H, Chernyavskiy T, Lu L, et al. Multiple Pathways to Resistance to ABT-378 Observed by In vitro Selection. 6th Conference on Retroviruses and Opportunistic Infections, Chicago, IL, USA 31 Jan � 4 Feb 1999. Absract 117.
Mayers, DL, Baxter JD, Wentworth DN, et al. The impact of drug resistance mutations in plasma virus of patients failing on protease inhibitor-containing HAART regimens on subsequent virological response to the next HAART regimen: results of the CPCRA 046 (GART). Antivir Ther 1999; 4 (Suppl 1):51.
Zolopa A, Shafer R, Warford A, et al. HIV genotypic predictors of antiviral response to saquinavir (SQV)/ritonavir (RTV) therapy in patients who have failed prior protease inhibitors (PIs): A clinical cohort study. 12th World AIDS Conference, Geneva, Switzerland, 28 June � 3 Jul 1998. Abstract 32287.
Tebas P, Patick AK, Kane EM, et al. Virologic responses to a ritonavir--saquinavir-containing regimen in patients who had previously failed nelfinavir. AIDS 1999; 13:F23�28.
Zolopa A, Tebas P, Gallant J et al. The efficacy of ritonavir (RTV)/saquinavir (SQV) antiretroviral therapy (ART) in patients who failed nelfinavir (NFV):a multi-center clinical cohort study. 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, USA, 26�29 September 1999. Abstract 2065.
UK Collaborative Group on Monitoring the Transmission of HIV Drug Resistance. Analysis of prevalence of HIV-1 drug resistance in primary infections in the United Kingdom. BMJ 2001; 322:1074�1075.
Little SJ, Holte S, Routy JP, et al. Antiretroviral resistance and response to initial therapy among recently HIV-infected subjects in North America. Antiv Ther 2001 6 (Suppl 1):21.
Little SJ. Is transmitted drug resistance in HIV on the rise? BMJ 2001; 322:1074�1075.
Duwe S, Brunn M, Altmann D, et al. Frequency of genotypic and phenotypic drug-resistant HIV-1 among therapy-naive patients of the German Seroconverter Study. J Acquir Immune Defic Syndr 2001; 26:266�273.
Hirsch M, Brun-Vezinet F, D�Aquila R, et al. Antiretroviral drug resistance testing in adult HIV-1 infection. Recommendations of an International AIDS Society-USA Panel. JAMA 2000; 283:2417�26.
Durant J, Clevenbergh P, Halfon P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomized controlled trial. Lancet 1999; 353:2195�2199.
Baxter JD, Mayers DL, Wentworth DN, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 2000; 14:F83�F93.
Cohen C, Hunt S, Sension M, et al. Phenotypic resistance testing significantly improves response to therapy:a randomized trial (VIRA3001). XIII International AIDS Conference, Durban, South Africa, 9�14 July 2000. Abstract ThPpB1433.
Meynard JL, Rosenthal J, Cameron M, et al. Impact of treatment guided by phenotypic or genotypic resistance tests on the response to antiretroviral therapy: a randomized trial (NARVAL, ANRS 088). Antivir Ther 2000; 5 (Suppl 3):67�68.
Tural C, Ruiz L, Holtzer C, et al. Utility of HIV genotyping and clinical expert advice - the Havanna Trial. Antivir Ther 2001; 6:S31�S32.
Call S, Westfall A, Cloud G, et al. Predictive value of HIV phenotypic susceptibility testing in a clinical cohort. 39th Interscience Conference on Antimicrobial Agents and Chemotherapy; September 26�29, 1999, San Francisco, USA. Abstract LB-17.
DeGruttola V, Dix L, D'Aquila R, et al. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antiviral Ther 2000; 5:41�48.
Hirsch MS, Conway B, D'Aquila RT, et al. Antiretroviral drug resistance testing in adults with HIV infection. JAMA.1998; 279:1984�1991.
The EuroGuidelines Group for HIV Resistance. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. Lancet 2001; 15:309�320.
Paredes R, Puig T, Arno A, et al. High-dose saquinavir plus ritonavir: long-term efficacy in HIV-positive protease inhibitor-experienced patients and predictors of virologic response. J Acquir Immune Defic Syndr 1999; 22:132�138.
Durant J, Clevenbergh P, Garraffo R, et al. Importance of protease inhibitor plasma levels in HIV-infected patients treated with genotypic-guided therapy: pharmacological data from the VIRADAPT Study. AIDS 2000; 14:1333�1339.
Kakuda TN; Page LM; Anderson PL, et al. Pharmacological basis for concentration-controlled therapy with zidovudine, lamivudine, and indinavir. Antimicrob Agents Chemother 2001; 45:236�242.
Top
|
|
|
|
|
|


